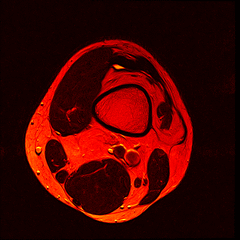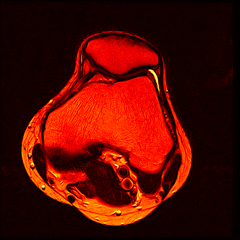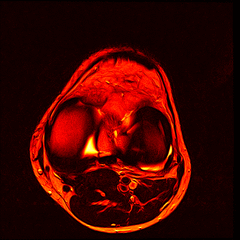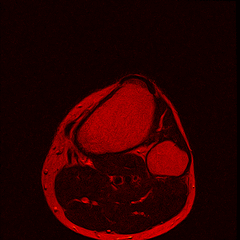
Over the past couple of years, I’ve had 2 MRI scans of my left knee as part of ongoing treatment/diagnosis for chronic pain. When discussing my treatment with the consultant, I was intrigued by the images produced by the scanner - detailed images of the inside of my knee from several angles. And thanks to the Data Protection Act, the hospital has to provide me with the data from thescans if I ask for it. So, one quick query at the MRI department, I handed over £10 (The nominal fee to prevent frivolous requests) and walked away with a CD containing the DICOM image data from both my scans.
Ok, so I have the data but what can I do with it? Surely I’ll need some proprietary medical imaging software to view it with? Well on the CD was a windows based viewer program but as a Linux user it’s no good to me. Thankfully because DICOM is a ubiquitous standard there are masses of tools available, including many open source offerings. I ended up trying both Ginkgocadx and Slicer3D. Both worked as expected but I ended up sticking with Ginkgocadx - it clicked with me more than Slicer, although that may just be because I couldn’t be bothered to read the Slicer tutorials. Both applications let you view, manipulate and edit the image sequences and they can produce 3Dmodels from the scan data as well, although I haven’t explored that option properly.
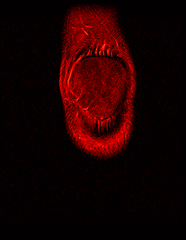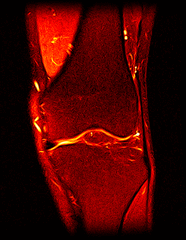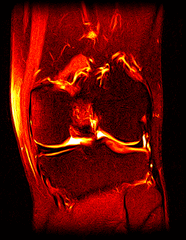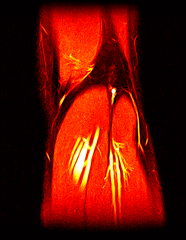
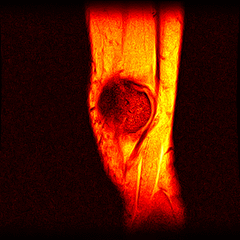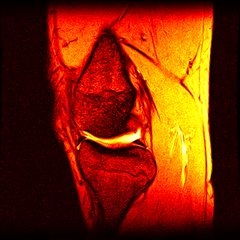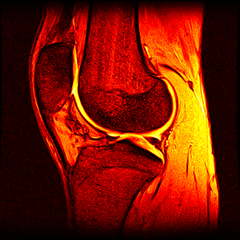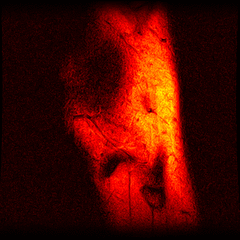
So far I’ve just colourised and converted the image data to make it easier to read and then produced the following pointless (yet strangely engrossing) animated gifs:
Bones, muscles, tendons and veins in glorious tango-vision!
These images came from the most recent scan, I’ve yet to convert the ones from before my arthroscopy but I’ll do that soon and then see if I can spot the difference.
If you fancy having a go yourself but haven’t had an MRI scan, then drop me a line and I’ll send you an anonymised version of my data.